Biological Processes and Interactions
Processes in Molecular Genetics: Between Genotype and Phenotype
MINDS ON
Some Key Terms
Look for these terms in this Activity. You should practice using them as you compose your responses to the upcoming tasks.
Realize, though, that this list is not exhaustive so you should look for opportunities to include other relevant terminology related to molecular genetics.
| tagging | radioisotope | semi-conservative |
| nucleotide | polymerase | helicase |
| primase | topoisomerase | ribosome |
| spliceosome | exon | intron |
| A-site | P-site | E-site |
| tRNA | mRNA | rRNA |
| primer | Okazaki fragment | codon |
| anticodon | transformation | replication |
| transcription | translation |
Tracking Biological Molecules

by Franklin, R., 1952.
Look back to your graphic organizer from Unit 1, Activity 6 on Cellular Structures, to see how different imaging technologies can be used to visualize parts of cells. Larger structures, like the nucleus, cell membrane and cell wall, can be seen using a light microscope. Individual components within these larger structures in addition to smaller organelles can be seen using electron microscopy. And the shapes of molecules, like proteins, ribosomes and nucleic acids, can be interpreted from X-ray patterns of X-ray crystallography.
While X-ray crystallography is a useful tool, samples of the molecule being studied need to be prepared in a test tube, or in vitro, providing limited understanding of how the molecule behaves inside cells. Another method can be used instead to follow biological molecules in cells: tagging. In this method, part of the molecule, or even a single atom, is marked in a way that allows us to see yet allows the molecule to function as it normally would. A common method of tagging uses radioisotopes (definition:an unstable isotope of an element that emits radiation.) of the elements that are found in biological molecules. Radioisotopes are also used in medicine to view changes in the body on the molecular level. The radiation from the radioisotopes is detected and the location of the radioisotope indicates where the tagged molecule is. The image to the right, for example, shows radiolabeled dopamine receptors in the brain. Hotter colours, like yellow or red, indicate more radiation caused by the presence of more radiolabeled dopamine receptors.

The element nitrogen is found in proteins and nucleic acids but not carbohydrates or lipids. On the other hand, phosphorus is found in nucleic acids and lipids, but rarely in carbohydrates or proteins. As you watch the following video, think about how different elements can be used to tag different types of macromolecules. If a cell is tagged with a radioisotope of phosphorus and then its macromolecules are separated, which macromolecules would emit radiation?
 Why is shape important in Biology?
Why is shape important in Biology?
Shape 18. Many structures and components in the cell are made of biological compounds that contain phosphorus. Identify which structures and specific components in the cell can be radiolabeled with a radioisotope of phosphorus.
Nucleic Acid Structures
Look back to your graphic organizer for Unit 1, Activity 5 about Nucleic Acids, to see that there are 8 common nucleotides: four deoxyribonucleotides and four ribonucleotides. The use of different nucleotides in cells depended on how those nucleotides were put together. DNA typically forms into double helices of two strands of deoxyribonucleotide polymers. RNA molecules, on the other hand, have distinct roles as monomers, dinucleotides or polymers with specific shapes.
 Tic-Tac-Know
Tic-Tac-Know
Review your Comparison of Nucleic Acid Structures from Unit 1, Activity 5 to refresh your memory about some of the details of this important macromolecule.
|
ribonucleotide |
double helix |
tRNA |
|
phosphodiester bond |
hydrogen bond |
polymer |
|
mRNA |
deoxyribonucleotide |
genetic information |
ACTION
The Hereditary Molecule
We take it for granted these days that DNA is the molecule that carries our genes. (definition:a segment of DNA that is responsible for the physical and inheritable characteristics or phenotype of an organism.) It is so prevalent in popular culture that it’s hard to imagine a time when it was needed to be shown that DNA was the hereditary molecule. As recently as 100 years ago, when biochemists separated nucleic acids, they could find no reactions that these macromolecules performed. It was known that nucleic acids usually came with large amounts of protein. Since proteins were known to do a wide variety of biological processes it was assumed that anything to do with heredity must also involve proteins. You can see more about the early ideas and experiments about DNA in the animation of this interactive.

Before we begin to look at how scientific understanding of the role of DNA, it’s important to be aware that also at that time Biologists understood that pathogens like bacteria and viruses cause many diseases. Biologists used their knowledge of pathogens to help learn more about DNA. These bacteria have special adaptations to their cell membranes that help them to escape destruction by the immune system. Viruses work by attaching themselves to the cell membrane of their host cells and, in the case of lytic cycle viruses, the host cell bursts open releasing hundreds of viral particles. A group of viruses called bacteriophages were known at that time to infect bacteria. Pathogens seem to interact with hosts at every step in a food chain.
 DNA Timeline
DNA Timeline
Several interesting experiments began to change scientific understanding to our current model of DNA as the hereditary molecule.
As you explore the following experiments, create a timeline to organize the discoveries in chronological order. For each experiment, identify the species being studied, in addition to the results of the experiment. Ask yourself, how do the results change scientific understanding of the role of DNA?
DNA
Each of the scientists worked as part of collaborative teams. Head researchers would often have Ph. D. students working for them in order to speed up experimenting. They also participated in the broader scientific community by meeting like-minded colleagues and publishing their findings in peer-reviewed journals. Some matches between researchers would prove to be more productive and allow for quicker discoveries.
Organization of DNA
DNA is organized differently in prokaryotes and eukaryotes. Because prokaryotes lack a nucleus, their DNA is organized in a nuclear region within the cytoplasm. Their entire genome is organized in a circular chromosome. In order to save on space, the chromosome is twisted up on itself.

Some prokaryotes also have smaller circular pieces of DNA called plasmids, although these generally don’t carry essential genes.
In the video we can see that eukaryotes also have chromosomes that are compacted to save on space.
The genome in eukaryotes can be organized on one or many different chromosomes.
Because of these differences in DNA organization, the way that DNA is copied and used also differs. We will explore processes involving DNA later in this Activity.
In all organisms, the nucleotides in each strand are connected by phosphodiester bonds. Biochemists number the carbons in the sugar part of the nucleotide with an apostrophe called “prime”, so the phosphodiester bond is made from the dehydration synthesis between a hydroxyl on the 3’ carbon of deoxyribose and the phosphate group on the 5’ carbon. You can see how this works in this familiar interactive showing dehydration synthesis between two nucleotides.
 Quiz
Quiz
Check your understanding of joining nucleotides. Visit the Dallas Learning Cloud, scroll down the page until you see the following, then click on the Launch Activity bar.

The double helix structure of DNA was correctly identified by Watson, Crick, Wilkins and Franklin as having an antiparallel orientation.
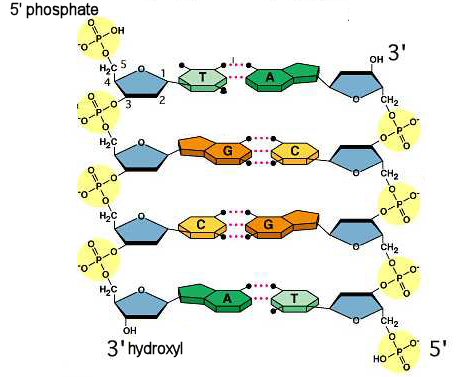
by Lesson Locker
It would be like writing one line in a paragraph rightside up and the next one upside down. It is only in this way that the nucleotides can be correctly orientated to make hydrogen bonds between the strands. We describe the two strands in terms of the functional group that could be used to make a new phosphodiester bond: either the 3’ hydroxyl or the 5’ phosphate. As we will see in this Activity, the orientation of nucleotides is important to the code in DNA, much like it is important to know which way to read a sentence in order to understand its meaning.
Look back to your graphic organizer from Unit 1, Activity 5 on Nucleic Acids, to see how each nucleotide makes hydrogen bonds with only one other nucleotide: A pairs with T and G pairs with C. We describe this base pairing as complementary base pairing because the second nucleotide completes the pair with the first.
Practice making complementary base pairs in this interactive.
ComplementaryBasePairs
Genotype to Phenotype
Soon after the double helix structure was published scientists began to work on discovering how DNA worked in cells. It was known that the proteins produced by cells is a heritable trait, and so Crick continued research into protein synthesis. With the understanding that DNA was the heritable molecule Crick proposed a theory he called the Central Dogma: DNA makes an intermediate molecule that is used to make protein.

by Dhorspool used under CC BY
Early work hypothesized that this intermediate molecule was made of RNA.
Three different RNA molecules play an important role in translation. As you explore each type of RNA at the Genetic Science Learning Centre, look to see how each of them interact with one another.
tRNA and rRNA are found only in the cytoplasm. mRNA is found in both the nucleus of eukaryotes, where DNA is, and the cytoplasm, where protein synthesis occurs. Today we call transcription the process of making mRNA molecules in order to make the primary structure of proteins. The process of protein synthesis is called translation.
Both transcription and translation consist of three steps: initiation, elongation, and termination. As we explore these two processes you will need to organize the details of the three steps.
 Processes in the Central Dogma
Processes in the Central Dogma
At the end of this Activity you will be presenting the steps of the processes in the Central Dogma as a narrative. A narrative describes steps connected together like in a story. The narrative should include details about initiation, elongation, and termination for each process. Watch one of the two following videos or explore the interactive to learn more about transcription and translation. Your narrative should include these details and concepts:
- location in the cell or organelle where these processes occur,
- the main enzymes and substrates for each process,
- details about initiation, elongation, and termination of each process.
Be sure to add more details as you work through this activity.
Transcription, in its simplest form, involves only one enzyme, RNA polymerase, and ribonucleotides as substrates to build the copied RNA strand. There are two sections in the DNA sequence that help RNA polymerase to identify where to initiate transcription and where to terminate it. A section of DNA rich in A and T nucleotides is the initiation site called the promoter. From there, RNA polymerase moves in the 3’ to 5’ direction reading the DNA and building the RNA strand.
Only one of the two strands is used to make RNA. The strand that is read in the 3’ to 5’ direction is called the template, or coding, strand. The other strand is not read so it's the non-template, or non-coding, strand. The hydrogen bonds are separated between the DNA strands so that the non-template strand is unzipped from the template strand. Just like between DNA strands in the double helix, the RNA is built so that it is antiparallel to the DNA template. This means that the RNA strand is synthesized in the 5’ to 3’ direction. Transcription ends at a sequence in the DNA called the terminator.

by Open Text BC
In prokaryotes, transcription occurs in the nuclear region of the cytoplasm. In eukaryotes, however, transcription occurs in the nucleus. The RNA strand produced by RNA polymerase is called pre-mRNA at this point because it needs to be modified before it moves through pores in the nuclear membrane into the cytoplasm.
The cytoplasm contains enzymes that are designed to hydrolyze DNA and RNA molecules as part of the cell’s natural protection from the genetic information from pathogens. RNAase enzymes can begin to digest RNA from either the 5’ or 3’ end. In order to protect the cell’s own RNA from these enzymes, the pre-RNA is modified with a modified nucleotide “cap” on the 5’ end and a long chain of adenosine nucleotides, or polyadenylate tail, on the 3’ end. The cap changes the shape of the RNA molecule so it can’t fit the active site of RNAase. The polyadenylate tail serves to protect the important information within the code of the RNA from RNAase by saturating their active sites with expendable, non-coding, RNA.
In eukaryotes, RNA is modified a third way in the nucleus. The way that genes are organized is that the information containing the code for the gene is broken up with sections of nucleotides which do not code for the gene. The section of the gene containing the code is called an exon with the in between sections that do not code for the gene are called introns. Special enzymes called spliceosomes work to remove introns. Similar to ribosomes, spliceosomes are made of RNA combined with proteins. For the purpose of this course, we do not need to focus on the details of this process.

by Open Text BC
Once the cap and polyadenylate tail are added, and the introns are spliced out, the finished mRNA is transported to the cytoplasm.
Complete this interactive to simulate the transcription of a gene in a eukaryote. Real genes are much longer, with longer introns between exons. You may find it useful to use good Independent Work skills (definition:I can use class time appropriately to complete tasks.) and your summary of the Processes of the Central Dogma from your Portfolio as you work to transcribe this gene.
RNABasePairs
 How do small steps lead to changes?
How do small steps lead to changes?
Changes 17. Predict what would happen in a cell with a shortage of ribonucleotides. How would that change the process of transcription?
After transcription mRNA binds to a ribosome to start translation. In prokaryotes, because both transcription and translation occur in the cytoplasm, it is common for these two processes to be coupled. Ribosomes and RNA polymerase are located close together near the nuclear region. Eukaryotes, on the other hand, separate the two processes with only translation occurring in the cytoplasm. Furthermore, ribosomes in eukaryotes are sometimes free in the cytoplasm or attached on the membrane of the rough endoplasmic reticulum.
In both eukaryotes and prokaryotes the ribosome is made up of two subunits that combine together to make a functional ribosome, similar to how certain proteins function in a quaternary structure made of tertiary structures connected together. The smaller of the two subunits binds to the cap on the 5’ end of the mRNA and begins moving towards the 3’ direction in search of its initiation sequence. Ribosomes look at three-nucleotide long blocks called codons. For most organisms, the same codon is used to initiate translation, similar to how a capital letter is used to start a sentence in most languages.
Each of the 20 amino acids are coded through the 64 codons. Special enzymes in the cell match the correct amino acid to the tRNA that corresponds to its correct codon. In this way the DNA is translated to protein using a codon “dictionary.”
Once a start codon is found, a tRNA binds to the mRNA. A three-nucleotide sequence on the tRNA, called the anticodon, is complementary to the codon on the mRNA. The larger ribosomal subunit then connects to the small subunit-mRNA-tRNA complex. Now the ribosome is complete and can catalyze the synthesis of a polypeptide chain.
The ribosome has three sites that are important for elongation in translation: the E-site, the P-site and the A-site. The mRNA is found in all three sites. The tRNA for the start codon during initiation is bound to the mRNA at the P-site. New tRNAs are added to the A-site but only if their anticodon is complementary to the codon in the mRNA in the A-site. Sometimes, to simplify how we explain the process of translation the E-site is not mentioned.
 Processes in the Central Dogma
Processes in the Central Dogma
As you explore the role of the P-site and A-site in one of the two interactives below, be sure to add good details to your summary of the Processes in the Central Dogma. Identify where the polymerization reaction occurs and in which direction the ribosome moves along the mRNA. How does translation terminate?

Proteins made in the rough endoplasmic reticulum are sent in vesicles to the Golgi apparatus to undergo further modifications and proper tertiary structure formatting. In this way the genotype coded in the DNA is converted to a phenotype that is caused by the proteins that leave the Golgi apparatus.
Use the following interactive to view the translation of mRNA.

 How do small steps lead to changes?
How do small steps lead to changes?
Changes 18. Predict what would happen in a cell if tRNA were charged so that any amino acid could bind to any tRNA? What about if no amino acids were attached to any tRNAs? How would these change the process of translation?
Genetic Continuity
Also after the double helix structure was published scientists began to work on discovering how DNA was copied in cells. There were three main hypotheses for how this might happen.

by unknown
In conservative replication the parent (definition:the original DNA strand.) DNA double helix is used to make an exact copy of itself but it is not changed in the process. This is similar to how a photocopier works. In dispersive replication the parent DNA is broken up into smaller bits that are later put back together. Some parts of the new double helices are made of the parent strands in between new sections of DNA. In semi-conservative replication the parent DNA double helix separated into single strands. Each single strand is used to make a complementary daughter (definition:the new DNA strand) strand. The new double helixes are made of a parent strand and a daughter strand.
Matthew Meselson and Franklin Stahl tested this hypothesis using E. coli bacteria grown in nutrients composed of an isotope of nitrogen. At the start of the experiment the nitrogen in the nitrogenous bases of the bacterial DNA is tagged with heavier N-15. See how the DNA made after the first and second generation of replication change in their weight after the bacteria are moved to ordinary light N-14 nutrients.

 How does understanding change?
How does understanding change?
Understanding 28. In an elegant experiment the results are so clear that conclusions can be easily made from them. Explain which specific details from Meselson’s and Stahl’s results are the best evidence that DNA is replicated semi-conservatively.
The process of replication is similar in a lot of ways to transcription. Again there are three main steps to this process: initiation, elongation and termination. There are a few more enzymes and proteins involved in replication which isn’t surprising given that in replication two strands are copied.
 Processes of the Central Dogma
Processes of the Central Dogma
You can add information about replication to your summary of the Processes of the Central Dogma. Explore the following interactive to get an overview of what is happening in each step.
Replication
Termination of replication occurs when replication forks meet and all the DNA has been copied.
In prokaryotes, there is usually just one origin of replication that helicase binds to.

by Catherinea228 used under CC BY
As the helicase moves around the chromosome it opens up the double helix to create a replication fork that exposes the nucleotides to enzymes. Replication ends when the two replication forks meet.
In eukaryotes, there are usually multiple origins of replication along each chromosome because the genome is often significantly larger than in prokaryotes.
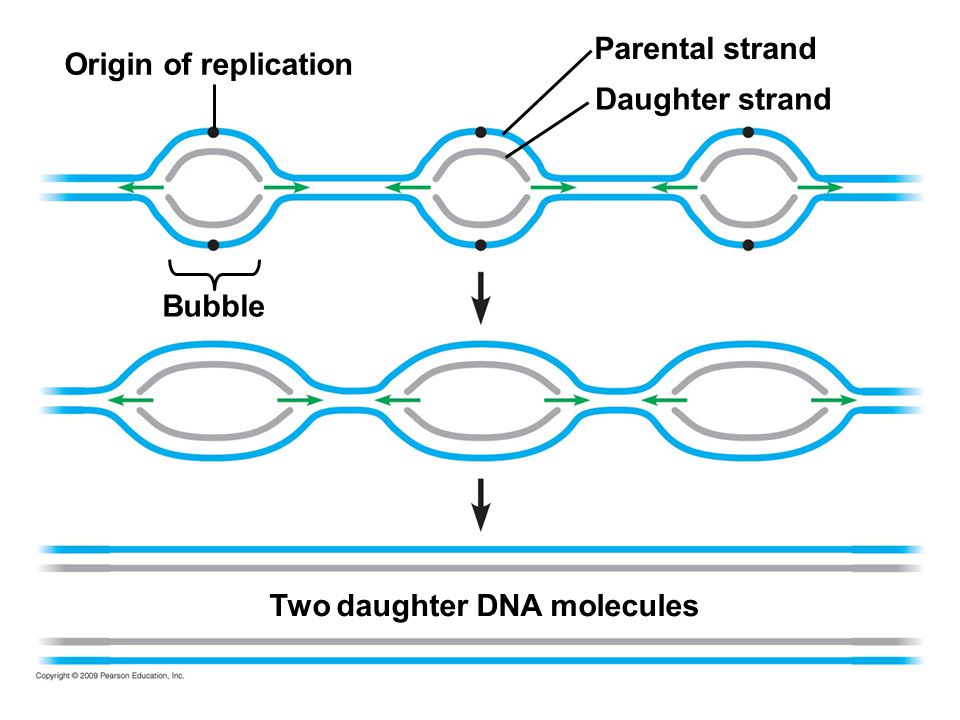
by Pearson Publishing
Again, replication ends when two replication forks meet.
In transcription the role of separating DNA strands and making RNA are both performed by RNA polymerase. On the other hand, in replication these roles are separated. To prevent the nucleotides from reannealing after being separated by helicase single stranded binding proteins attach to the single stranded portion of the DNA until it is copied.
 How do small steps lead to changes?
How do small steps lead to changes?
Changes 19. Predict what would happen in a cell if single stranded binding proteins had a change in shape so they could no longer bind to single stranded DNA? How would that change the process of replication?
Just like in transcription, the main polymerases in replication, primase and DNA polymerase III, read the DNA template in the 3’ to 5’ direction. New nucleotides are added in the 5’ to 3’ direction. However, in replication both strands of DNA are copied so that adds a few extra steps. One strand is synthesized quickly and is called the leading strand. The other strand is made slowly so it’s called the lagging strand.
 Processes of the Central Dogma
Processes of the Central Dogma
As you explore either the interactive or the video look to see why the two strands of DNA are synthesized differently. Add these details to your summary of Processes of the Central Dogma.
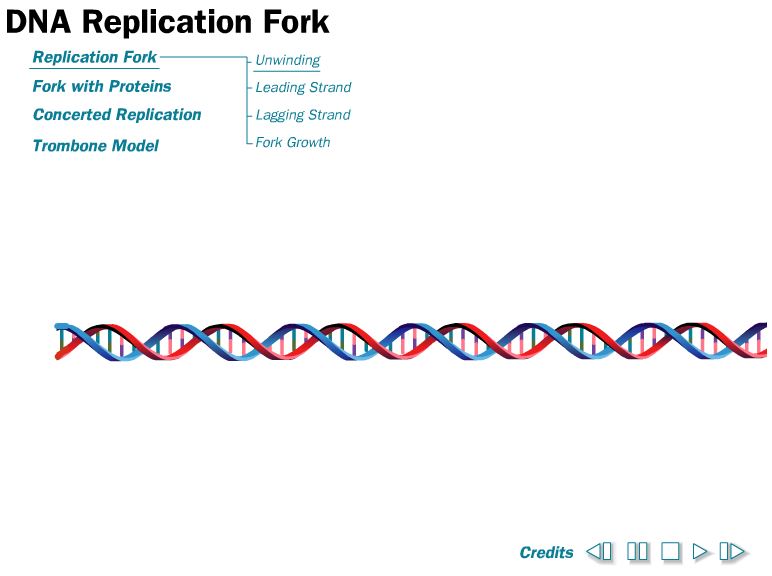
 Why is shape important in Biology?
Why is shape important in Biology?
Shape 19. How does the orientation of DNA strands explain why Okazaki fragments are made on the leading strand but not the lagging strand? You may find it useful to think of your answer in terms of the direction that the enzymes move during replication.
Telomeres

There are over 20,000 genes in the human genome across each of our 23 pairs of chromosomes. The genes are organized in the chromosomes away from the telomere ends or the centromere towards the middle. The centromere serves as the attachment point for spindle fibres during cell division. Telomeres, on the other hand, don’t have a direct function in the day-to-day processes in the cell. Their length, however, is important and has been correlated to a variety of health indicators ranging from aging to cancer.
The length of telomeres is maintained by a special polymerase called telomerase. Changes in the ability of telomerase can lead to changes in the rest of the cell.
 Why is shape important in Biology?
Why is shape important in Biology?
Search for a change in telomerase activity related to aging or a disease. Describe how the length of telomeres is changed. How does the change in telomerase activity relate to a symptom of the disorder or aging.
 Processes of the Central Dogma
Processes of the Central Dogma
After decades of research into the steps in replication there is a sophisticated understanding of this process. The following video is a computer animation that combines evidence from several imaging technologies. The real process happens much faster than what we see here.
Does your summary include these details and concepts?
- The location in the cell or organelle where these processes occur,
- When each process occurs compared to one another as part of a sequence of steps,
- The main enzymes and substrates for each process,
- Details about initiation, elongation, and termination of each process.

 Processes of the Central Dogma
Processes of the Central Dogma
What did you learn about the processes of the Central Dogma?
3 - Three things I learned about how DNA is utilized by cells:
2 - Two other things I found interesting about the processes of the Central Dogma:
1 - One question I still have:
CONSOLIDATION
Summary
- the structure of DNA is important in understanding how it is used;
- DNA is transcribed into mRNA that is then translated in protein synthesis;
- DNA is copied semi-conservatively.
 Processes of the Central Dogma
Processes of the Central Dogma
Your task is to describe how a specific gene is replicated and used to make its protein.
Choose an important gene to human cells. There are over 20,000 genes in our genome so to help you narrow it down you can think of an enzyme or protein we have studied earlier in the course. You will need to find and describe each of the following details:
- on which chromosome the gene is located,
- how the chromosome is opened up to allow for replication or translation,
- how DNA is replicated,
- how a gene is transcribed,
- how a gene is translated,
- what modifications the protein undergoes after translation,
- how the gene product is used in the cell.
Present your information in a narrative style that describes a series of steps. Think of how the sequence of the processes can be organized into an interesting storyline. You may find you can compose an interesting narrative by making an infographic, an animation, or a comic strip.
Your narrative should include these details and concepts:
- location in the cell or organelle where these processes occur,
- the main enzymes and substrates for each process in the Central Dogma,
- details about initiation, elongation, and termination of each process in the Central Dogma,
- how gene product is relevant to human cell functions.